Roses are red,
Violets are blue,
If I were evolution,
I’d select you.
Author
Adam Kane: kanead[at]tcd.ie
Photo credit
Floridapfe

Trinity College Dublin, Ecology and Evolution
The Natural History Museum in London is one of my favourite places. The majesty and beauty of the building’s design is a fitting exterior to house the truly stunning collections within.
The new Treasures exhibition displays just 22 of the museum’s most prized possessions. It’s a special opportunity to see valued and varied treasures such as the type specimen of the earliest known bird, Archaeopteryx, Darwin’s pigeons and the Iguanodon teeth which sparked the discovery of the dinosaurs all lined up together. The stories behind the origin and significance of each of the treasures are fascinating.
Although not one of the most famous objects, the Emperor Penguin’s egg was my favourite item. The egg is beautiful in itself but its real value as a treasure lies in the story behind its collection. It is one of just 3 intact specimens which were collected by Captain Scott’s ill-fated Antarctic expedition between 1910 and 1913. In this centenary anniversary year, Scott’s quest to reach the South Pole remains one of the most inspiring examples of human endeavour. Before the age of GPS, insulated clothes or re-heatable “expedition food”, Scott’s crew ventured into the heart of the frozen continent. As if striving to reach the pole wasn’t enough of a challenge in itself, the expedition also had the aim of collecting as much scientific data about Antarctica as possible. Their chief scientist Edward Wilson wanted to examine penguin embryos for evidence of an evolutionary link between birds and reptiles. However, Wilson was part of the team who perished with Scott on their return journey from the Pole. The specialist embryologist who was going to study the eggs died in the First World War and by the time results from studying the eggs were published in 1934, the evolutionary recapitulation theory on which the egg-study was based was outdated.
The story behind this egg certainly puts the trials of modern scientific research into perspective. While the rate of new scientific discoveries shows no sign of slowing, delving into the finer details of the inner workings of the cell or the evolution of drug-resistant bacteria doesn’t hold quite the same level of physical adventure as that which was represented by Scott’s expedition. I’m not for one instant suggesting that I long for a more dangerous yet adventurous age or that modern fieldwork is not without its trials and difficulties. I just don’t know of any current research projects which can match Scott’s story in terms of raw human endeavour into the most unknown, dangerous and inhospitable conditions imaginable. It was a treat to see such a precious and fragile reminder of past scientific endeavour on display.
The treasures exhibition is a stunning collection of prized objects and should be treated as a site of pilgrimage for anyone even remotely interested in evolution or the natural world. Most of the specimens are unique but fortunately an example of one treasure, the Great Auk can be seen in the TCD Zoology museum. So that’s one ticked off the list for Trinity – though I’ve a feeling that it might take just a little while for us to match the rest of London’s collection …
Author
Sive Finlay: sfinlay[at]tcd.ie
Photo credit
wikimedia commons
February 5th marked the 70th anniversary of the first lecture of what was later to become Schrödinger’s highly influential book ‘What is life’.
While Schrödinger may be more popularised by his infamous zombie cat, it was his thinking with regards to how life can live with the laws of physics that have allowed him to transcend that major divide between the physics and biological communities.
Schrödinger’s genius insight was to see life as a system behaving and constrained by the second law of thermodynamics, in particular describing the probable nature of a hereditary crystal, which would later lead scientists including Shannon and Weaver to the discovery of DNA and the genetic code.
However what makes the story of this work more fascinating, in particular to me as I have been lucky enough to get the opportunity to give a (very) small talk in the same lecture theatre were Schrödinger gave his lectures 70 years ago, is the story of how an Austrian physicist ended up in the capital of Ireland writing some of the most important work in biology of the last century.
Schrödinger, an Austrian, fled Germany in 1933 due to his dislike of the Nazi’s anti-Semitism and became a fellow in Oxford, during which time he received a Nobel Prize with Paul Dirac. However things turned sour with Oxford due to his less then monogamous approach to the opposite sex, and the lack of acceptance towards living with his wife and mistress lead him to leave for Princeton. These problems followed him there and eventual he found himself back in Austria in 1936. The occupation of Austria by the Germans in 1938 led him to again flee, this time to Italy. But in the same year Eamon de Valera, Ireland’s Taoiseach (Prime Minister) at the time, personally invited him to Dublin were he spent the next 17 years.
It was here in Trinity College Dublin that he delivered his lecture series which were to inspire both Watson and Crick to search for the genetic molecule and which has recently seen some increased popularity due to its use by Brian Cox in his latest series What is Life. However this work came about I like to think that “What is life” found its way to Ireland through Nazis and polygamy, a story surely of the calibre for the Discovery channel, if only it had some sharks.
Author
Kevin Healy: healyk[at]tcd.ie
Photo credit
wikimedia commons
“A lie can travel halfway round the world before the truth has got its boots on” (Mark Twain, attributed).
In my previous post I gave some background on intelligent design, the theme of a talk I recently attended by Dr Alistair Noble. This time, I’ll try and address his claims.
It is easy to say something that is not true. It is not always so easy to explain why it is not true. Such is my problem here. I can summarise Dr Noble’s arguments into a few sentences, but it takes paragraphs to explain why they are wrong. Here goes!
His argument centered around DNA. Dr Noble’s background in chemistry, specifically in trying to artificially synthesise chemicals, showed him how difficult it was to make even simple molecules. He explained his problems with DNA and used two specific examples to illustrate his argument: the bacterial flagellum and cytochrome C. His arguments were essentially:
The design argument can be easily refuted. Apparent design does not mean actual design. Humans are extremely good at seeing things where they do not exist, like shapes in clouds and Jesus on burnt toast. This is a well-known psychological phenomena called paradolia and can lead us to see design where none exists.
The second claim requires a bit more care. DNA, the bacterial flagellum and Cytochrome C are all highly complex and could not have evolved by chance. In fact, as Dr Noble so carefully illustrated, Cytochrome C would have taken longer than the lifetime of the universe to arise by chance. So if they did not arise by chance then they must have arisen by design, surely? Well, no.
This conclusion can only be made if you have a deep misunderstanding of evolution. At a very basic level random mutations occur which may be beneficial, neutral, or detrimental to an individual. Then natural selection ‘selects’ those mutations which are beneficial and ‘rejects’ those that the detrimental. Small changes over long timescales lead to big changes, mutations can build on each other and can be co-opted to other functions. The bacterial flagellum is a perfect example, with studies showing how molecules were co-opted from other functions to form the flagella. At no point was there a useless proto-flagellum.
ID proponents, including Dr Noble, focus on the random aspect of evolution but completely ignore the selection part, which is arguably the more important aspect. If there were no natural selection then their claims would be valid, but its presence provides a beautifully simple explanation of how complex molecules, complex biological components, and even complex organisms could arise.
Next time, my review of the talk.
Author
Sarah Hearne: hearnes[at]tcd.ie
Photo credit
wikimedia commons
It’s the year 2050. Several billion more humans occupy the world, and species translocations are by now the norm to mitigate against increased urban sprawl, climatic instability and a sea level now a third of a metre higher. In spite of unprecedented demands on the natural environment, governments have slowly developed capacity for conservation of wilderness and semi-natural habitat. Beyond this even, with the vast majority of the human race by now living in cities and the continued trend of rural land abandonment; restoration ecology has come to the fore at entire landscape and regional scales. The concept of ‘rewilding’ is debated openly amongst politicians and the public – no longer the mere theoretical exercise of academics. The monetary value of ecosystem services is also by now a very real and tangible concept within economic circles, embedded within highly developed metrics such as green-GDP. Despite such positive developments, however, problematic legacies of the past remain. Intensification of agriculture has been unrelenting globally, notwithstanding inroads into adoption of agroecosystem approaches. A transition to truly renewable energy sources is still incomplete and of utmost urgency. One of the most critical questions of all most likely still looms – have we yet done enough to put a cap in the peak of this, the sixth great mass-extinction of life on the planet?
And so, it is within this future and none-the-less challenging world we find the modern ecologist and biodiversity practitioner at work.
What kind of new and useful technologies may exist to help tackle such problems and challenges of the not so distant future? It is interesting to deliberate on one low-tech tool in particular (the so-called bread and butter of biodiversity), which has been with us already for centuries – and that is the humble species checklist. Specifically we take a look at the Flora – and although coverage here is rather phyto-centric, it should be easy to draw equivalents to all forms of taxa, without (too) much stretch of the imagination.
So what is a Flora in the traditional sense, why is this changing, and how will the Flora of the Future look and function? To briefly tackle these first two questions, a Flora is primarily a list of plant biodiversity (either with or without diagnostic characters and keys) within a specified geographic range, be it local, national or at larger scales. Outside of this basic function there are the ‘added-extras’, which may include notes on distribution, ecology, synonymy, conservation status and even ethnobotanical use. Often the assemblage of national-level Floras has proven quite a mammoth task; logistically challenging, fraught with funding difficulties, and above all time-consuming – with efforts spanning over several decades for particularly biodiverse countries. This is all very well, and such traditional Floras have and will continue to serve as invaluable tools. In this modern age, however, change is called for to tackle some common short-comings of the Flora. A considerable amount of valuable information collected by taxonomists and other experts in the production process is typically lost, never making its way into the public realm – and when such publications can easily run to over 20 volumes, it is clear to see the major constraints involved. Another key drawback is the sheer speed at which redundancy can occur. Even before the final volume of a Flora is published, taxa (species/genera/families) covered within the first volumes may have long been ripe for new taxonomic treatment.
The revolution in how biological information is collected, stored and disseminated is already greatly influencing the Flora. One of the most recently initiated national-level projects is the Flora of Nepal project, for which advances in biodiversity informatics have permeated the entire process from preparation to publication. Although the Flora of Nepal will still be published in printed format, a (if not the) main focus will be an E-Flora freely accessible online, which will also greatly expand the availability of information assembled by experts. A simple yet very significant feature will be the ease of portability of numerous volumes to the field in digital format. Though perhaps most critically, the Flora of Nepal will be maintained and updated to reflect new findings – creating for the first time, in essence, an evolving Flora.
Before we really begin to speculate on the form and function of our Flora of the Future, we must first take a look to the current cutting edge of biodiversity informatics. In what must be one of the most significant advances in decades, the cooperative development of the Global Biodiversity Information Facility (GBIF) by many governments and organisations has promoted and facilitated the “mobilization, access, discovery and use of information about the occurrence of organisms”. This centralized repository of earth’s biodiversity is fast set to reach one billion indexed records within a few years from now, fed from diverse sources ranging from individuals to national biodiversity data centres. It is difficult to envisage how the Flora of the Future could in any plausible way side-step such a global network. Whereas floras have traditionally featured a top-down, expert driven synthesis – the Flora of the Future will also no doubt integrate the emergent trend of bottom-up assembly of knowledge – a good example of which is currently purveyed by the Encyclopaedia of Life.
Let’s get back now to our future ecologists and biodiversity practitioners, and take a little look in as they go about conducting their fieldwork. No matter what habitat or location they study in worldwide, they will each possess a small handheld device connected to the Flora of the Future. Automation of species identification by means of this device will have removed a large bottleneck in their work – leaving ecologists to focus on actual ecology. No longer will they be bound to a particular geographic territory due to limited floristic familiarity – we will witness a complete opening of boundaries, and greater migration of ‘western’ ecologists to the frontline of areas of global biodiversity importance.
But just how exactly could such a device work? A potential basis could feature a combination of machine-learning morphometrics and DNA barcoding – two presently very promising tools. For the former, development of algorithms for auto-identification of plant species is already well underway (see for example the Leafsnap mobile app). These function much like facial recognition technology, and through input of a digital scan/photo can pinpoint unique morphological characteristics required for successful classification. A key aspect of machine learning is removal of subjectivity by conversion of shapes into numerical descriptions – no need for argument any longer on just how ‘subglobose’ a feature is; the ball is already in motion towards a predictive and integrative taxonomy. Upon scanning a specimen in the field, an image will be broken down into key morphometric characteristics, and referenced against a large central database within the Flora of the Future. The Flora will prioritize this procedure by first referencing against species known to occur within a certain radius from where the user currently stands (a useful feature in itself!). The ecologist, on the spot, may learn that the specimen has a confirmed match, and proceed to download key local statistics of importance. On the other hand, this specimen may in fact represent an extension of the species known distributional range. The finding, however, of no known match in the database could spell discovery of a new species, whereas a positive match with notably low morphometric agreement may indicate new subspecific taxa or otherwise interesting findings (for which DNA barcoding could be employed for further verification in both cases).
Regardless of outcome, the above three scenarios will have allowed for a real-time and in situ solution to identification of species. The exact significance of this process will not only lie in the freeing up of both ecologists’ and taxonomists’ resources, but in the real-time flagging of new discoveries. As it stands, it is expected that discovery of remaining undescribed plant species will be an incredibly inefficient process (given that 50% of the world’s plant species have been discovered by only 2% of plant collectors), despite the vast number of these thought to exist. A recent study examining the exact inefficiency of the production chain from collection to publication uncovered that “on average, more than two decades pass between the first collection of samples of a new species and the publication of the species’ description in scientific literature”. In other words, a specimen of a new species has physically passed through the hands of many people before the simple ‘discovery’ (perhaps after many, many years in a herbarium) that it is something new to science. In this sense, an important function of the Flora of the Future will be instant recognition (perhaps even while standing in the field!) of a new discovery as just that – which can drastically reduce this presently overblown timeframe and waste of resources.
Getting back to the future for now, we see our biodiversity practitioners and ecologists as key players in the advancement of ecological as well as taxonomic discovery, with a highly efficient yet passive ability for discovery embedded within the commonplace tools they use, as they go about their work. With an entirely streamlined approach to field research, and identification no longer a daunting prospect in the study and documentation of biodiversity, we will eventually see the peak of mass extinction pass, looking back behind us. The challenges of tomorrow are no doubt great, and a renewed vigour for the taxonomic process will be critical for progress on these fronts. The Flora of the Future will for the first time sew a seamless line between ecologists and taxonomy; the essential currency of biodiversity.
Author
Paul Egan: eganp5[at]tcd.ie
Photo Credit
Paul Egan
Cats eh? You either love them or you hate them it seems. Well the latest research published in Nature by Loss et al. (2013) will give those who hate them plenty more reason to do so. While those who love their cats may just sit that little bit less comfortably next to their feline companions.
Let me start by making a few things clear. Cats are predators, they are an invasive species which have been introduced to islands all over the world, by man. In many places domestic cats have become feral, i.e. reverted to living in the wild, which has led to huge increases in their numbers in some places, which can have a devastating effect on indigenous wildlife populations. For a more detailed and somewhat depressing example of where this has occurred read about Stephens Island in New Zealand. Famously a lighthouse keeper’s cat had been blamed for the extinction of an entire species on this island though it seems that reports may have been somewhat exaggerated in this case.
The report in Nature is more scientifically robust than urban legends about a lighthouse keeper’s cat though. Figure 1 below shows just how devastating domestic cats can be on local wildlife populations. The graphs show the estimates of predation by domestic cats on (a) birds and (b) small mammals.
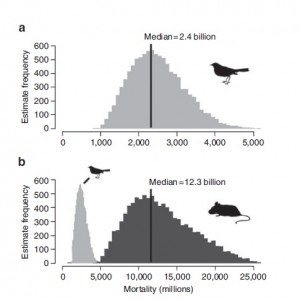
The numbers are startling, an estimated 2.4 billion birds are killed by cats every year in the US and 12.3 billion mammals. Incredible numbers I’m sure you will agree, there is however a caveat; only 31% and 11% for birds and mammals respectively are caused by what the writers class as “owned cats”, cats which are regularly kept indoors and well fed. While the majority of the mortality is thought to be caused by free roaming “unowned” cats. Incidentally there has recently been some debate about wind farms and their impact on local bird populations but this excellent blog and another recent Nature piece put the numbers into perspective in terms of other anthropogenic causes of bird deaths.
As those responsible for the domestication and introduction of cats, we can’t lay all the blame at the feet of our feline friends. First of all we need to somehow effectively manage the populations of feral and “unowned” cats and while this has been attempted with the Trap-Neuter-Return movement, it has been viewed as a response based on regarding feral cats as part of the native fauna rather than the invasive aliens that they are , therefore largely unscientific and ineffective. Secondly pet owners can take several measures of their own, neuter or spay your cat while keeping your cat indoors at night can vastly reduce their impact on local wildlife.
Author
Keith McMahon: mcmahok[at]tcd.ie
Photo credit
wikimedia commons
At a recent session of NERD club, our weekly research group meeting for the Networks in Ecology/Evolution Research Dynamic, we discussed academic CVs. Four academic staff members (including myself) showed their CVs to the group and discussed what was in them. This was interesting because we all had such varied opinions! I thought I’d write a short blog post to highlight some of our main agreements and disagreements. Continue reading “Academic CVs: Dos, don’ts and maybes”
We the blog declare that a month of games will commence from tomorrow. The aim is to achieve the most hits for a blog post in a day. The prize will be worth that of a King’s Ransom and will be revealed in good time. Cry havoc, and let slip the blogs of war!
Scribe
Adam Kane: kanead[at]tcd.ie
Photo credit
wikimedia commons
Trinity College Theological Society recently held a talk by Dr Alistair Noble titled ‘A Scientific Case for Intelligent Design’ which I attended as, possibly, the only biologist in the room. It was a fascinating, if deeply frustrating, experience. Before I get into the details of the talk, a brief explanation of intelligent design may be necessary. . .
Intelligent design (ID) is the ‘theory’ that certain features of the universe, including life, are best explained by invoking a creator. I put ‘theory’ in quotes because in a scientific theory is a very particular beast. It must have both explanatory and predictive powers. For example, the theory of evolution by natural selection explains how life evolved and can also be used to make predications about life that can be tested. The ‘theory’ of intelligent design has little explanatory power (“the designer did it”) and makes no predictions. As such, it is held with little esteem within the scientific community.
Outside the scientific community, however, there are some who hold ID in very high esteem. They think that it is a credible scientific theory and there have been many attempts, particularly in the U.S., to have ID taught in schools as a counter to evolution. This is deeply worrying to those who care about scientific literacy but has to be tackled carefully.
The reason for such caution is that ID is most loudly promoted by religious groups who feel that the theory of evolution is anathema to their beliefs and as such must be countered. In the past they countered with Creationism, but in recent years they have tried to remove the explicit religious overtones of Creationism, removing God, replacing him with an unspecified ‘designer’ and calling the new theory ‘intelligent design’. Thus the debate around ID is not just a scientific debate but is also a religious debate involving deeply held personal beliefs.
I hold the opinion that your personal beliefs are yours, and are no concern of mine, but when you try and mess with science, well, that’s another story! I went to the talk as I was curious to hear the scientific evidence for ID. Would it persuade me that there was a case for ID? . . .
Author
hearnes[at]tcd.ie
Photo credit
wikimedia commons
The media is all abuzz about the Carter Centre’s recent announcement that 542 cases of guinea worm infection were reported in 2012. That is a remarkable achievement, considering that 3.5million cases where the reported when the Carter Centre began their eradication programme in 1986. The guinea worm (Dracunculus medinensis) is a particularly gruesome parasitic nematode that causes painful and debilitating disease. It is one species no one will be too sorry to see go. Well no one except the folks at the (tongue in cheek) Save the Guinea worm Foundation.
Perversely, considering our track record of causing extinctions, actually trying to get rid of a species can be extremely difficult. Targeted eradication of disease in humans has been successful only once before, with small pox. That required a massive and expensive vaccination programme and it is unlikely that the mandatory aspect of the vaccines would be tolerated today. However, helminths are a different beastie altogether. Helminths (parasitic worms) differ from pathogens in that, with a few exceptions, they don’t multiply within human hosts or have direct transmission. Helminths require a period of passage through the environment, either as infectious eggs or through other intermediate hosts. The guinea worm life cycle involves water fleas (Cyclopidae) as intermediate hosts. Water containing infected water fleas is drunk and the parasites are released. After about a year of maturation, females emerge via a painful skin blister, which erupts on contact with water, releasing thousands of larvae ready to continue the cycle.
The peculiarities of the life cycle meant the eradication programme was successful, not though vaccination or medication, but through changing people’s behaviour in the key areas of transmission and infection. To prevent infection people were taught about the need to filter drinking water, particularly standing water where cyclops abound. The burning sensation caused by the female worm emerging meant people often cooled the blister in a nearby pond, usually the same the one that supplied drinking water. By educating about the link between this behaviour and infected ponds, transmission of the larval stages was successfully reduced.
Of course, various other aspects of the guinea worm life cycle played a part. Cyclops is a relative large (1mm) so filtering material could be made and supplied cheaply. They are also immobile; once an infection is eradicated from an area it is easier to keep it out than in diseases like malaria. Unlike helminths that release eggs and larvae through the intestinal tract, people shedding guinea worm infectious stages are much more likely to be identified quickly.
One important factor influencing the success of small pox eradication was that the virus had no hosts other than humans. There is no wildlife reservoir from which the disease may re-emerge. Guinea worms on the other hand have been found in cats, dogs and cattle, though none appear to act as a reservoir for human infection. It may, therefore, be more correct to speak of elimination of human guinea worm infections rather than total eradication of the species. Save The Guinea Worm Foundation will be pleased.
Author
loxtonk[at]
Photo credit
wikimedia commons